. I graduated from
the Department of
Pharmaceutical Chemistry in March of 1997. Despite departing the
bay area, I still retain active collaborations with Peter Kollman and
the AMBER developers and
love to visit whenever I can...
. I graduated from
the Department of
Pharmaceutical Chemistry in March of 1997. Despite departing the
bay area, I still retain active collaborations with Peter Kollman and
the AMBER developers and
love to visit whenever I can...
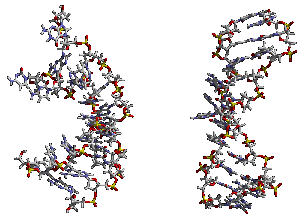
My thesis research in the Kollman group involved the realistic simulation of nucleic acids in solution. Tremendous amounts of computer time, both locally and at (what was at the time) a NSF sponsored supercomputer center, the Pittsburgh Supercomputing Center, were applied. Initially, the results we saw with the Cornell et al. force field and a cutoff were less than satisfactory (see the picture to the right), however, thanks to a collaboration with Tom Darden and assimulation of the particle mesh Ewald code, we were able to realistically and reliably simulate a variety of nucleic acid systems. This was aided by parallelization of the PME code by Mike Crowley and co-workers at PSC. See my resulting publications for more specific details... Highlights were the observation of spontaneous A-DNA to B-DNA transitions of DNA in solution, stabilization of A-DNA in mixed water/ethanol solution, and B-DNA to A-DNA transitions when hexaammine cobalt(III) ions were bound to GpG regions in the major groove of DNA.