Gallery
Molecular dynamics force-fields
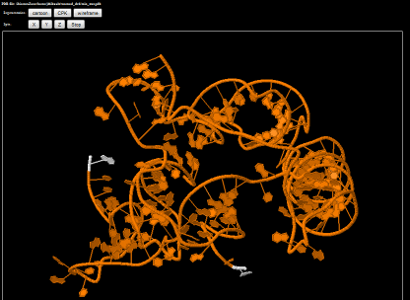
Reliable representation of a large variety of nucleic acid systems (DNA and RNA)
Drug design
 |
P450 enzyme study
Molecular dynamics refined structures of drug-metabolizing P450 enzymes are modified with quantum mechanics based parameters
for the catalytic heme to identify structural contributions to competing reaction mechanisms that can either safely eliminate
compounds or produce toxic metabolites.
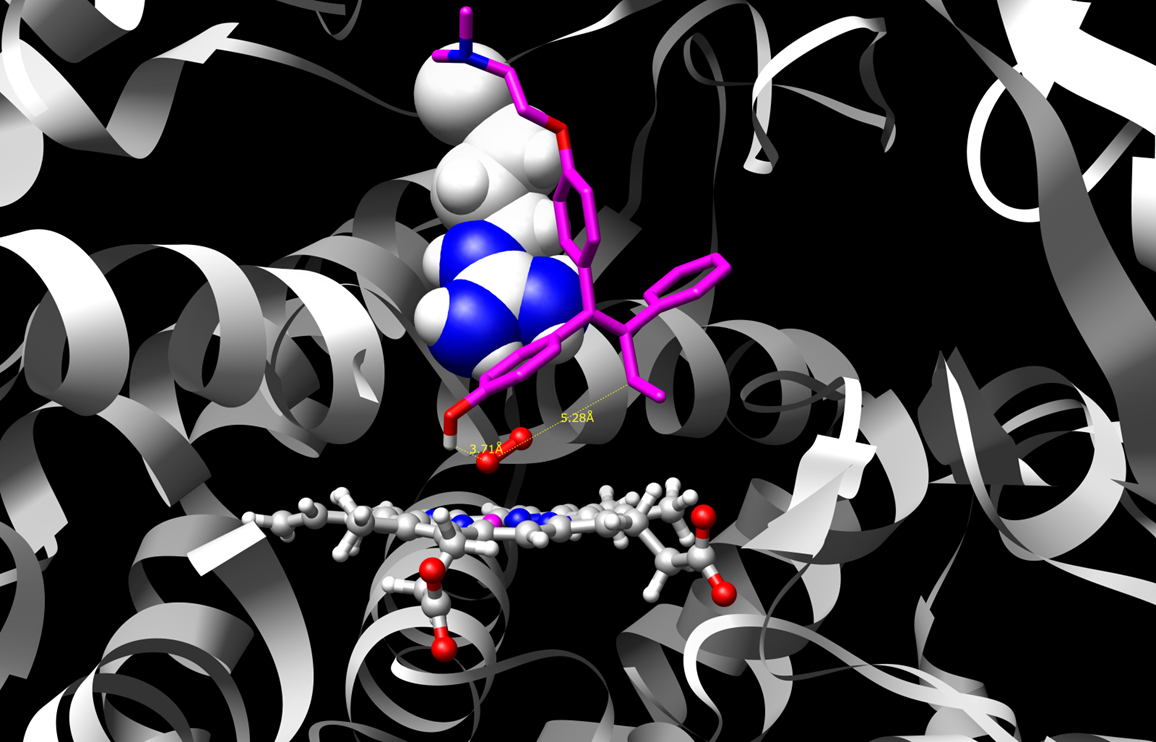
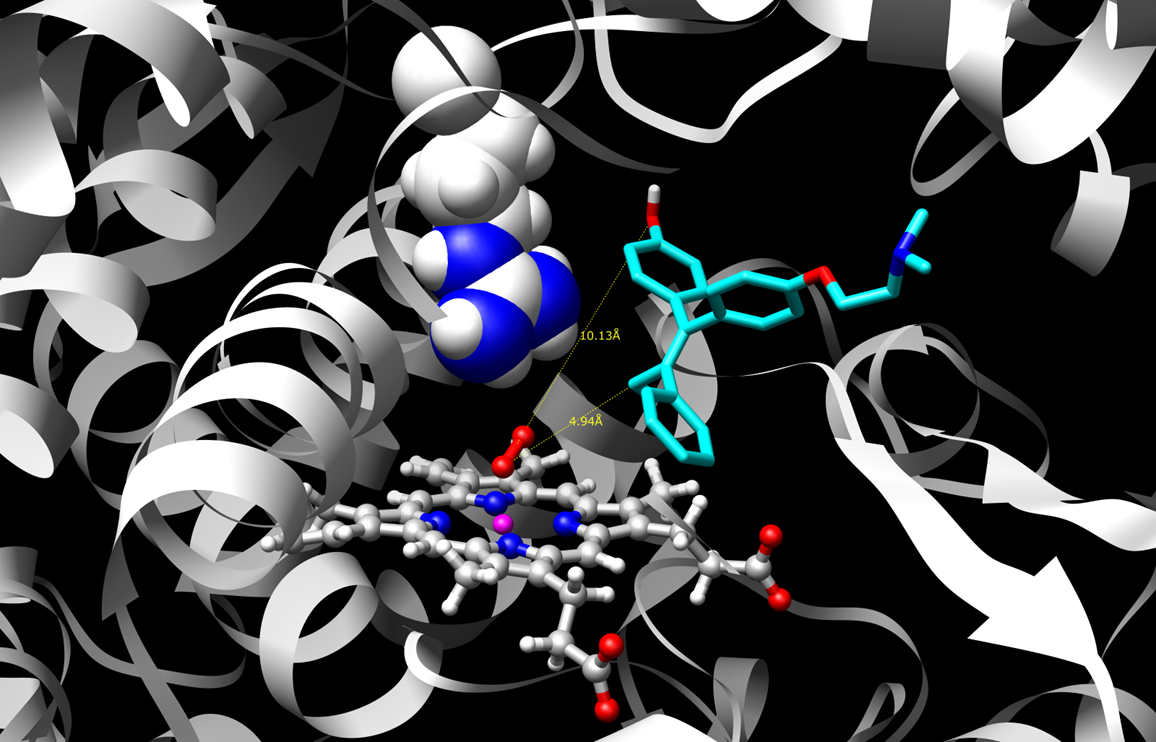
Two such poses or binding modes are shown for a sample substrate, 4-hydroxy-tamoxifen.
Figure 1 shows the substrate in a binding mode that is supportive of dehydrogenation. This reaction mechanism produces
reactive quinone methide species. Figure 2 shows a pose that supports hydroxylation which leads to safe drug clearance.
|
 |
Data mining and dissemination
 |
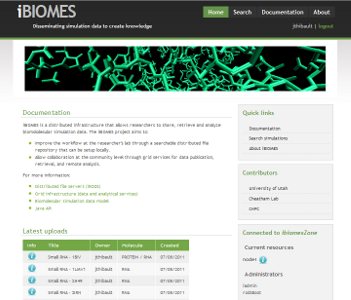
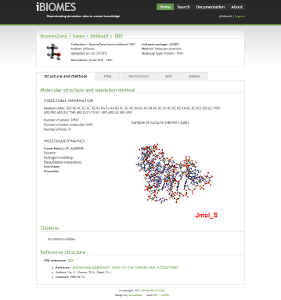
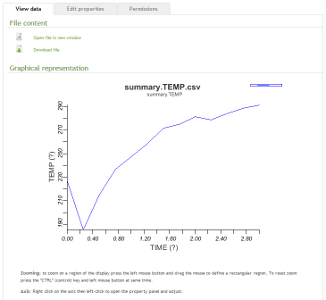
The iBIOMES project
The aim of the iBIOMES infrastructure is to allow data sharing and collaboration between researchers working in biomolecular simulation.
For more information please visit: http://ibiomes.chpc.utah.edu
|