Research
Focus
 The people in
our lab use and develop molecular dynamics, free energy
simulation, and trajectory analysis methodologies in applications
aimed at better understanding biomolecular structure, dynamics and
interactions. A strong focus of our funded efforts centers on the
reliable representation of nucleic acid systems (DNA and RNA) in
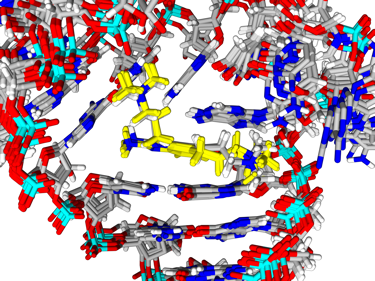
solution. For example, we helped solved the NMR structure of the
drug-bound Hepatitis C virus IRES structure shown on the left. Based
on this (and related structures), we can now apply CADD methods and
simulation to better
understand and design potential new Hepatitis C therapeutics. In
addition, large efforts are underway to better characterize RNA
structure and force fields through simulation of a large number of
commonly observed RNA structural motifs and a large variety of NMR and
crystal structures. We are also involved with international
collaborative efforts to understand DNA structure, for example through
the ABC
consortium and long simulations of
DNA...
The people in
our lab use and develop molecular dynamics, free energy
simulation, and trajectory analysis methodologies in applications
aimed at better understanding biomolecular structure, dynamics and
interactions. A strong focus of our funded efforts centers on the
reliable representation of nucleic acid systems (DNA and RNA) in
solution. For example, we helped solved the NMR structure of the
drug-bound Hepatitis C virus IRES structure shown on the left. Based
on this (and related structures), we can now apply CADD methods and
simulation to better
understand and design potential new Hepatitis C therapeutics. In
addition, large efforts are underway to better characterize RNA
structure and force fields through simulation of a large number of
commonly observed RNA structural motifs and a large variety of NMR and
crystal structures. We are also involved with international
collaborative efforts to understand DNA structure, for example through
the ABC
consortium and long simulations of
DNA...
Critical to reliable representation of the structure, dynamics and interactions is not only trying to simulation the biomolecules in their native solution environment but to also both critically assess and validate the simulation results with experiment. Our group focuses on both brute-force and enhanced sampling/ensemble-based simulation using available high performance computational resources at the University of Utah (www.chpc.utah.edu) and elsewhere. Outside resources include large allocations of computer time from Blue Waters, XSEDE (www.xsede.org), on the Anton machine at PSC and from other sources. With these resources we also are able to expose and overcome limitations in the methods and force fields...
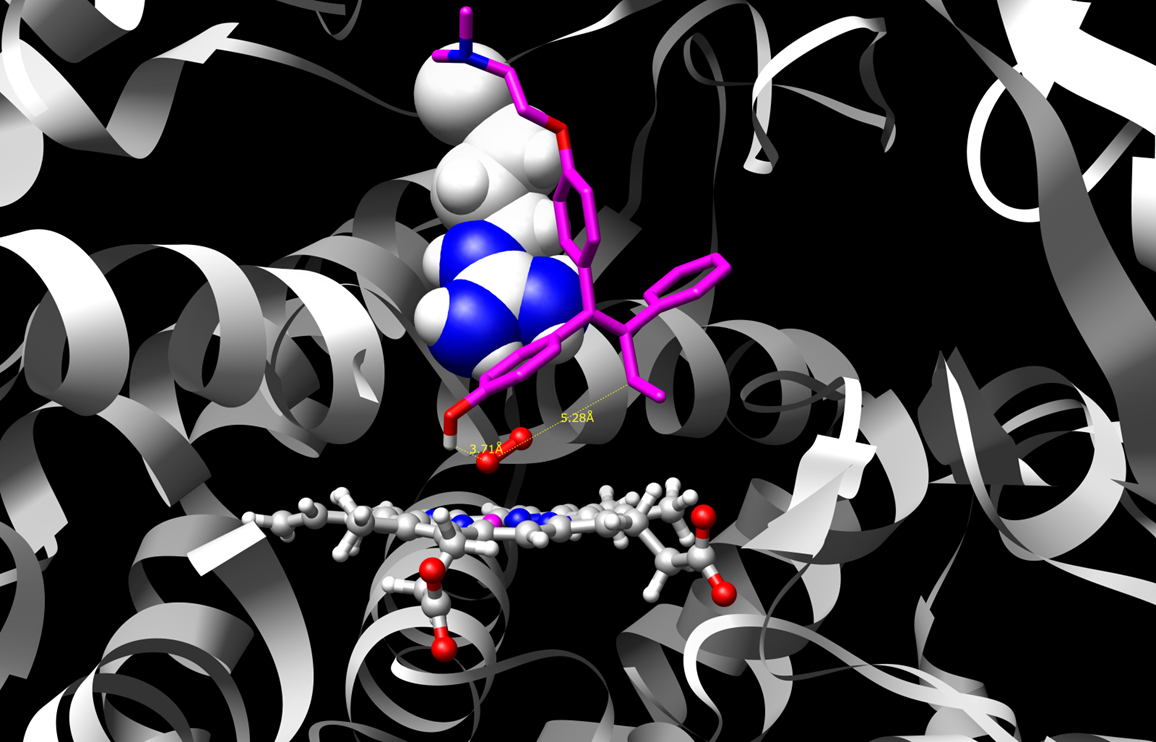 Beyond nucleic acids, we are also interested in various coiled-coils,
enzymes and cytochrome P450's. A key emphasis is on improving
stability or understanding how ligands alter receptor structure upon
binding. Also, in addition to continued development of the
ptraj/cpptraj tools within the AmberTools suite for analysis of MD
trajectories, we are exploring methods to mine more information from
the simulation data and means to more broadly disseminate the MD
results.
Beyond nucleic acids, we are also interested in various coiled-coils,
enzymes and cytochrome P450's. A key emphasis is on improving
stability or understanding how ligands alter receptor structure upon
binding. Also, in addition to continued development of the
ptraj/cpptraj tools within the AmberTools suite for analysis of MD
trajectories, we are exploring methods to mine more information from
the simulation data and means to more broadly disseminate the MD
results.
Although our primary development and simulation engine is
AMBER, we also use and have experience
with CHARMM, NAMD, and other
programs.
Software
Follow the links to the software.
Grants
Research in the lab is currently funded by various research grants, including:
NIH RO1-GM081411: "Biomolecular simulation for the end-stage refinement of nucleic acid structure".
(2/01/08-1/31/14) PI: Cheatham.
This core R-01 funding supports research into the development of better force fields for simulation of RNA, attempts various means
to assess and validate the performance of MD simulation as applied to RNA, and seeks to explore means to more broadly disseminate MD simulation data.
Here is the NIH
reporter link.
Currently this is in no-cost-extension.
NIH RO1-GM098102: "RNA-ligand interactions: Simulation and experiment".
(9/30/11-8/31/15) M-PIs: Kathleen Hall (WUSTL), Cheatham and Carlos Simmerling (Stony Brook U).
This core R-01 funding supports research into the development of better force fields for simulation of RNA, attempts various means
to assess and validate the performance of MD simulation as applied to RNA, and seeks to explore means to more broadly disseminate MD simulation data.
Here is the NIH reporter link
NSF CHE-1266307: "CDS&E: Tools to facilitate deeper data
analysis, exploration, management, and sharing of ensembles of
molecular dynamics trajectory data".
(10/01/13-9/30/15) PI: Cheatham
Further development of the iBIOMES environment for sharing and
disseminating MD simulation data and also tools for deeper analysis
(CPPTRAJ).
NSF ACI-1341034: "CC-NIE Integration: Science slices
converting network research innovation into enhanced capability for
computational science and engineering at the University of Utah".
(10/01/13-9/30/15) PI: Corbato, Co-PIs: Bolton, van der Merwe, Ricci, Cheatham
Deployment of a science DMZ network at the U of Utah.
NSF OCI-1035208: "PRAC - Hierarchical
molecular dynamics sampling for assessing pathways and free energies
of RNA catalysis, ligand binding, and conformational change".
(2/01/11-1/31/14) PI: Cheatham, Co-PIs: Simmerling (Stony Brook),
Roitberg (U Florida), and York (Rutgers).
This is a travel grant that provides training and preparation for
eventual usage of the "Blue Waters" supercomputer at NCSA. We are one
of ~36 groups that will be given access.
Here is the
link to the NSF awards database..
NIH R01-GM074249: "P450-mediated dehydrogenation mechanisms."
(1/01/11-12/31/15) PI: Gs Yost Co-PIs: Cheatham, Reilly
Experiment, simulation and docking probe the interaction of substrates
with P450 with a focus on dehydrogenation reactions.
Here is the
NIH reporter link
NSF Cyberinfrastructure partnership, LRAC MCA01S027 (yearly)
"Insight into biomolecular structure, dynamics, interactions and
energetics from simulation".
PI: Cheatham.
This is a large allocation of resources on the NSF supercomputer
centers to support research in the Cheatham lab.
Recently ended grants
NIH R01-GM079383: "AMBER force field consortium: A coherent biomolecular simulation platform."
(9/28/07-8/31/12) PI: Y. Duan (UC Davis) Co-PIs: Cheatham, C. Simmerling (Stony Brook U), R. Luo (UC Irvine), P. Cieplak (Burnham Inst), and J. Wang.
The consortium aims to improve the AMBER force fields; our role
focuses on polarizable force fields for nucleic acids.
Here is the
NIH reporter link.
Pittsburgh Supercomputing Center, PSCA00033P
"Molecular dynamics of DNA and protein-DNA complexes: A proposal
for obtaining microsecond trajectories using Anton".
PI: Cheatham.
Pittsburgh Supercomputing Center, PSCA00067P
"Development and testing of improved fixed-charge force fields for proteins".
PI: Case.


